Results
 The DiCOVA Track-1 Challenge (COVID-19 detection fromm cough sounds) received registrations from 85 teams, spread across the globe and coming from industry, academia and independent individuals. All these teams were sent the challenge datasets. Of these teams, 29 participated in evaluating their systems against the blind test set (233 audio files).
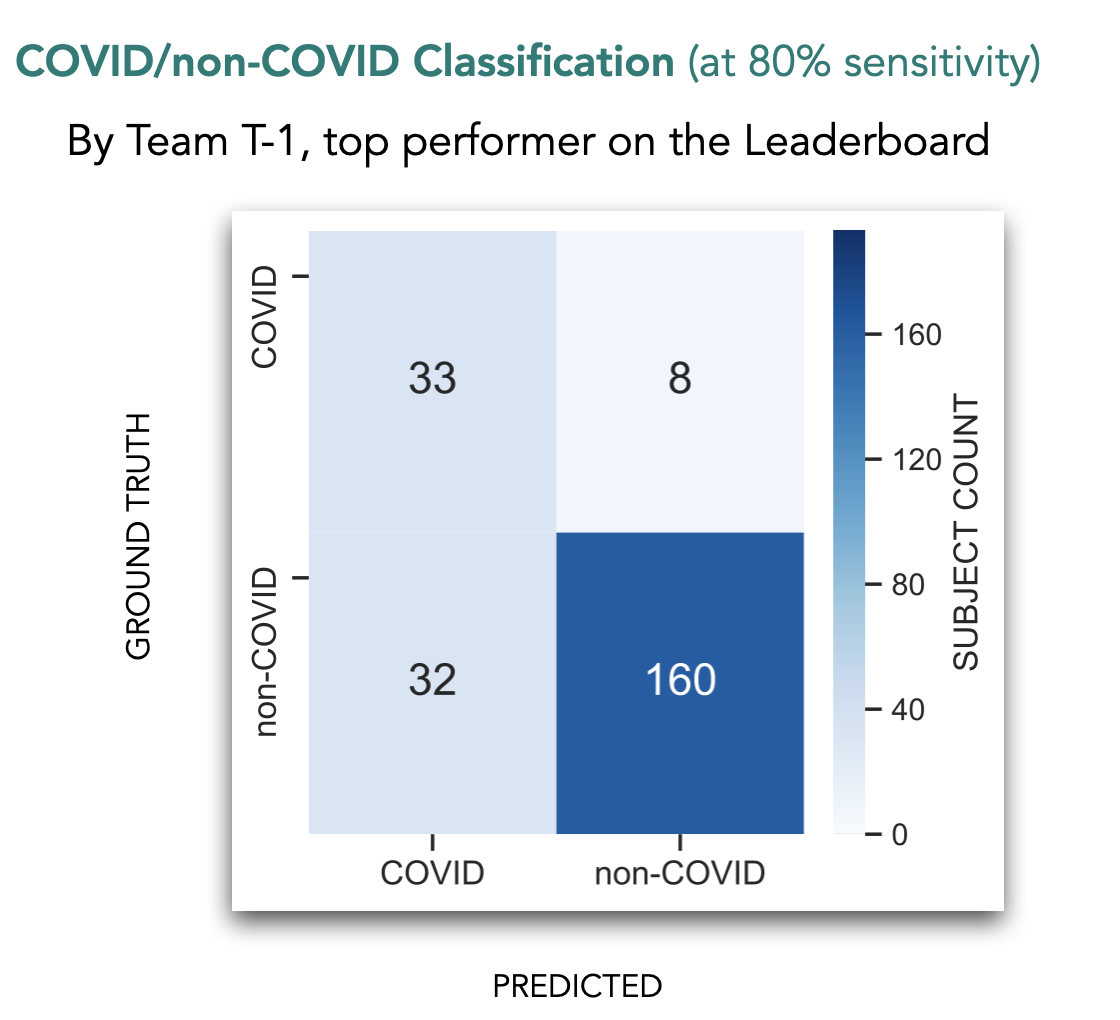
For this, a leaderboard was set up in Codalab and teams posted a COVID probability score for each test audio file. In response, they received the AUC score (area under the ROC curve) computed over the 233 test audio files. A high AUC (0-100%) implies better performance. Team T-1 posted an AUC of 87.04% and finished on top of the leaderboard. On the right you can see the classification performance of this team on the blind test set.
Below we illustrate a few of our observations on the activity seen on the leaderboard.
The DiCOVA Track-1 Challenge (COVID-19 detection fromm cough sounds) received registrations from 85 teams, spread across the globe and coming from industry, academia and independent individuals. All these teams were sent the challenge datasets. Of these teams, 29 participated in evaluating their systems against the blind test set (233 audio files).
For this, a leaderboard was set up in Codalab and teams posted a COVID probability score for each test audio file. In response, they received the AUC score (area under the ROC curve) computed over the 233 test audio files. A high AUC (0-100%) implies better performance. Team T-1 posted an AUC of 87.04% and finished on top of the leaderboard. On the right you can see the classification performance of this team on the blind test set.
Below we illustrate a few of our observations on the activity seen on the leaderboard.
The leaderboard saw participation from 29 teams.
Each team was given a maximum of 25 attempts to evaluate their system performance against the hidden blind test labels. The AUCs of many of these systems performed better than the baseline system.
There was a good diversity in kinds of features used by the teams. These features ranged from simple hand-crafted acoustic features (like, ZCR, energy) to advanced acoustic representations (embeddings) obtained using pre-trained DNNs.
The novelty of the task made teams also experiment with diverse kinds of classifiers.
The challenge task required handling class data imbalance. For this, several teams experimented with data augmentation (adding noise, reverberation, pitch shifting, etc., or cough files from other public datasets, like COUGHVID), and system fusion.
The best performance was posted by team T-1 with an AUC of 87.04%, significantly improving over the baseline system performance (69.85%). This performance was followed by two close competitors, team T-2 posting 85.43% AUC and team T-3 posting 85.35% AUC. It was wonderful to see nine teams scores above 80% AUC!
The evaluation was open for 22 days. In the initial days only a few teams evaluated their systems. As days passed, the leaderboard activity began to gain pace, and teams started improving their AUCs.
How did the best AUC on the leaderboard change over evaluation days?
Does more evaluation by a team imply a better AUC? There is some correlation :)!
How does the performance on the test set compare against the performance on the val set?
An important metric in evaluating a diagnosis tool is its specificity at some sensitivity. For the challenge we evaluated the specificity at 80% sensitivity. Below we show how different systems fared in this. The best specificity obtained was 83.33% by team T-1.
And finally, here are the ROCs of the 29 systems corresponding to the best system of each team.